4 Ways To Analyze Tissues By Flow Cytometry

Why are we interested in cytometry? What are its practical applications? Let’s start by defining cytometry as the measurement of cellular processes at the whole-cell level. This definition is useful because it includes not only flow cytometry, but any technique that measures at the level of the whole cell. Microscopy, for instance, is a great example of cytometry. But, why analyze tissues?
When flow cytometry is practiced, the cells are broken up. Therefore, any cellular interactions within the sample are also broken up. This includes cell-to-cell contact and virtually any information about the microenvironment. As we continue to discover, the microenvironment can play a dramatic role in cell development, influencing how cells grow and change.
Here are 4 ways to analyze tissues by flow cytometry…
1. Analyze with Multi-photon cytometry
Multi-photon microscopy doesn’t rely on traditional fluorescent excitation. In the traditional process, a single photon will excite the molecule. Following this, there is a fall back to the ground state along with the desired fluorescent emission. But in multi-photon cytometry, you can take advantage of two-photon excitation. Here, there are 2 photons — typically of higher wavelengths in the 700-1000nm range — that hit at the same time, providing enough energy to excite the fluorescent molecule. As with traditional excitation, a fall back to ground state ensues.

Figure 1: One vs two-photon excitation. In two-photon excitation, two lower-energy (higher wavelength) photons simultaneously excite the fluorochrome.
But, the real value of multi-photon microscopy lies in its allowance for deeper tissue penetration. It can actually be done along with live-cell imaging in the brain, heart, or certain other organs to show biological processes in real-time. A paper by Miller includes cell imaging results covering a 5-minute time-lapse. During this lapse, the movement of immune cells can be observed. In the images, you might notice that each cell moves just slightly differently. By using this process to measure cells in animal tissue, you’re able to see motion that is difficult to capture in static images, like serial sections or by using traditional fluorescent microscopy.

Figure 2: Results from multiphoton microscopy experiment. From Miller et al, (2003)
2. Analyze with Plate-based cytometry
In plate-based cytometry, cells are measured on a plate. A microwell plate, or something similar, will be ideal. This method works especially well for measuring adherence cells, or any cell that can be made to stick to the plate. The instrument used will be a Celigo, a plate-based cytometer. In Figure 3, the cells are plated in a very flat, defined tissue well. The wells are scanned using the Celigo, which allows for either a fractional scan or a whole-well scan as desired.

Figure 3: The Celigo process.
If a system allows for multiple fluorescent parameters, you can use a DNA dye like DAPI. Or, you can even try rhodamine or one of the far-red dyes. In Figure 4, there are 3 panels indicating the overlays of different staining processes. The first panel represents tubulin, phalloidin, and DAPI. In the second panel are cells labeled with phospho-ERK and Hoechst to label the nucleus. The third panel features GFP and mCherry. Just to recap, if you do have adhering cells, this is far better than pulling them off a plate and placing them in a traditional flow cytometer.
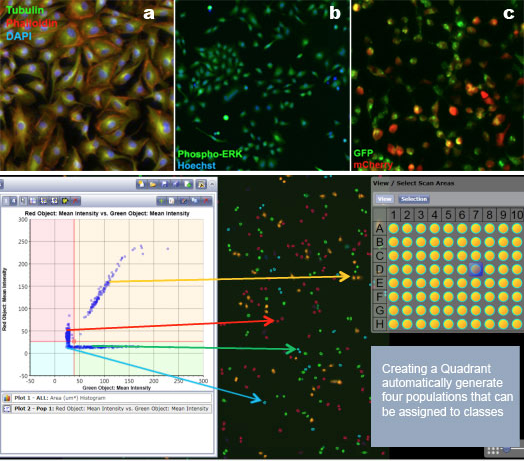
Figure 4: Typical Celigo data analysis using both image analysis and bivariate plots. Data from the Nexelom website.
3. Analyze by Imaging mass cytometry
Imaging mass cytometry has started to pick up some attention over the last few years. While the method itself has been around for a while, it only became popular when the CyTof corporation introduced new cytometry technology, including the Hyperion system. The Hyperion allows you to perform tissue-based mass cytometry. As seen in Figure 5, a sample is first prepared and labeled with antibodies. Here, you can use as many as 30 or 40 antibodies, which the CyTof device can measure.

Figure 5: Tissue mass cytometry process as illustrated by the Bodenmiller lab.
The tissue is laser-ablated, and during this process, the ions are thrown off. The resulting data can be collected using either traditional technology, or the newer options provided by CyTof. Then come processing and analysis, as usual. This method does leave you with subpar cellular resolution, but you can see whole-cell imaging and get a good look at whole-cell interaction. There will be about 30 parameters that can be observed for comparison and noting any changes.
The data in Figure 6 are from a review paper by Singh et al., showing different ways to use tissue cytometry. Imaging mass cytometry can be used to investigate things like germinal centers in lymph nodes, tonsils, or a wide range of tumor cells; just by watching the cell interactions. If you are using analytical software like SPADE or tSNE, you can produce a data plot and determine cell types and locations within your images.

Figure 6: Representative data from mass cytometry imaging, as described in Singh et al. (2017)
4. Chip cytometry
Chip cytometry, the product of German company Zellkraftwerk, is the final technique on this list. As shown in Figure 7, chip cytometry works by taking all the cells and putting them on a special chip. The cells are stained and imaged before fluorescence is neutralized. Then, the cells are stained with another marker, and this cycle is repeated many times over. On the right in Figure 7 is a 95-flex phenotyping plan developed by Zellkraftwerk. Their methodology permits 95 analytes simultaneously, which is beyond what even CyTof technology can manage.

Figure 7: A chip cytometry diagram from Zellkraftwerk showing the process (left) and a 95-plex panel (right). This technology can measure 2 to 3 times the number of targets as mass cytometry.
The power of chip cytometry is shown in Figure 8. Here, a labeled tonsil cell was examined with 32 markers to define the cells and explore differences due to inflammation of the tonsil. Shown in the top-left, traditional gating tools are available, with the added benefit of being able to go and look at the specific cell of interest. Furthermore, the other markers on this cell can be examined and a complete phenotype defined.

Figure 8: Chip cytometry data using inflamed tonsil cells and a 32-plex panel. Data from Zellkraftwerk.
When cells are treated to create the single-cell suspensions required for traditional flow cytometry, important information can be missed. Cell-cell contact, microenvironmental influences, and other factors can impact both cell growth and protein expression. Thus, having methods to explore these characteristics is a critical item in any researcher’s toolkit. The 4 methods described above offer different ways to obtain this information, and allow for a more complete understanding of the biology involved in processes like tumorigenesis, cell migration, tissue development, and more. So why measure in tissues? Because when flow cytometry is practiced, it becomes possible to glean a wealth of information about a microenvironment, and research continues pointing to the dramatic role a microenvironment can play in cellular development.
To learn more about the 4 Ways To Analyze Tissues By Flow Cytometry, and to get access to all of our advanced materials including 20 training videos, presentations, workbooks, and private group membership, get on the Flow Cytometry Mastery Class wait list.

ABOUT TIM BUSHNELL, PHD
Tim Bushnell holds a PhD in Biology from the Rensselaer Polytechnic Institute. He is a co-founder of—and didactic mind behind—ExCyte, the world’s leading flow cytometry training company, which organization boasts a veritable library of in-the-lab resources on sequencing, microscopy, and related topics in the life sciences.
More Written by Tim Bushnell, PhD












